Discovery Program Outcome Pathways

How can we incorporate human-based animal alternatives into drug discovery? Can we reduce animal testing and still improve program success? Making good decisions requires integrating information from many assays and experiments.
We have recently proposed a decision support framework, Discovery Program Outcome Pathways (DPOPs) to facilitate this process for drug discovery programs. DPOP is based on the Adverse Outcome Pathway (AOP) framework, a well-accepted structured knowledge representation used in chemical safety risk assessment (see also here and here). DPOPs and AOPs help organize information from diverse sources, at multiple levels of biological complexity, in order to facilitate decision making.
In the DPOP framework, assays are mapped onto a logical framework that connects molecular level information with clinical outcomes. The molecular initiating event (MIE) is usually the drug-target interaction (when known) and is connected to the outcome (some clinical measure) through mechanistically defined (or causal) intermediate steps, so-called key events. These intermediate key events include pathway-, cellular-, tissue- and organ-level processes. Results from target-based assays and phenotypic assays as well as more complex systems (e.g., organ chips, etc.) can be mapped onto different elements of the DPOP framework, as shown in the graphic below.
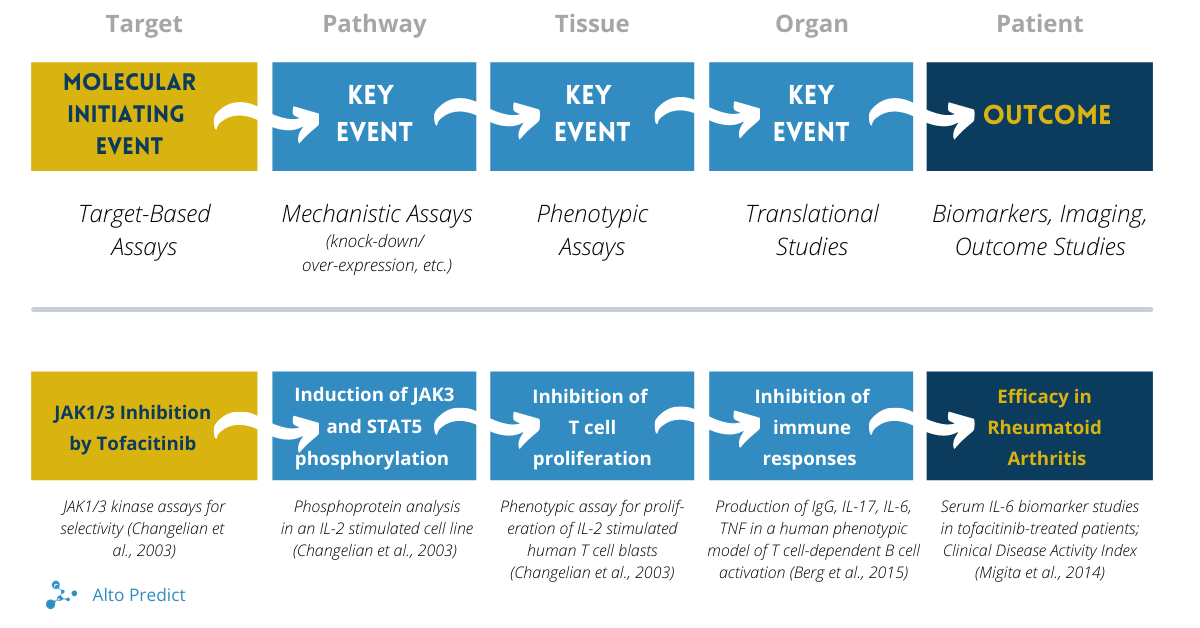 The Discovery Program Outcome Pathway Framework, adopted from Berg et al., 2021. The top panel shows the general framework. The bottom panel shows a specific example for the arthritis drug tofacitinib (Xeljanz®), a JAK kinase inhibitor, developed from assay data reported in Changelian et al., 2003, Berg & O’Mahony, 2015, and Migita et al., 2014.
The Discovery Program Outcome Pathway Framework, adopted from Berg et al., 2021. The top panel shows the general framework. The bottom panel shows a specific example for the arthritis drug tofacitinib (Xeljanz®), a JAK kinase inhibitor, developed from assay data reported in Changelian et al., 2003, Berg & O’Mahony, 2015, and Migita et al., 2014.
Example DPOP for Tofacitinib
As an example, program assays for the drug tofacitinib (as described in Changelian et al., 2003) can be mapped to the DPOP framework, shown in the lower section of the graphic). Tofacitinib (Xeljanz®) is a JAK kinase inhibitor approved in the US in 2012 to treat rheumatoid arthritis patients. The interaction of tofacitinib with JAK3 and to a lesser extent JAK1 and JAK2 kinases represents the MIE and was assessed using in vitro kinase enzyme assays. The induction of JAK3 and STAT5 phosphorylation represent a key event at the pathway level. This was assessed by measuring JAK3 mediated phosphorylation of itself as well as STAT5 in an IL-2 stimulated cell line assay. The next key event is inhibition of T cell proliferation and was measured in an assay using IL-2 stimulated human T cell blasts. The key event at the next higher level of biological organization is inhibition of immune responses. This was assessed using an assay system modeling T cell-dependent B cell activation where effects of tofacitinib on IgG, IL-17, IL-6 and TNF-alpha are measured (Berg and O’Mahony, 2015; O’Mahony et al., 2018). At the outcome level, biomarker studies have shown tofacitinib-dependent reduction in serum levels of IL-6 in patients with rheumatoid arthritis, supporting the clinical translation (Migita et al., 2014).
DPOP frameworks help us discover and confirm the causal connections between levels of biological organization which are crucial for developing new and better therapeutics. DPOP frameworks can be used to assess the strength of the chain-of-translatability, an important factor for drug discovery program success, and help identify gaps in knowledge. The use of DPOPs also help facilitate the incorporation of human non-animal alternatives into drug discovery decision making. This is a stated goal of the FDA in their recently published report, Advancing New Alternative Methodologies at FDA.
Look for more DPOP examples to come!
Images created in Canva by author.






