Are you ready for it? Technology readiness for in vitro NAMs

Faster adoption of alternatives to animals in biomedical research is a worthy goal for both scientific and ethical reasons.
Addressing key elements of technology readiness for cell-based alternatives (in vitro NAMs or New Approach Methodologies) will help get these tools out of research and into the hands of users.
In vitro NAMs or New Approach Methodologies are cell-based assays and platforms that provide practical models to help us understand how biological systems work, test hypotheses, and predict outcomes for specific questions. These tools are critical for advancing human-relevant science in biomedical research.
In vitro NAMs technologies are diverse, ranging from simple two-dimensional (2D) cell cultures to complex in vitro models (CIVMs), such as microphysiological systems (MPS) or organs-on-a-chip. They are complicated technologies and are not well understood by many potential users.
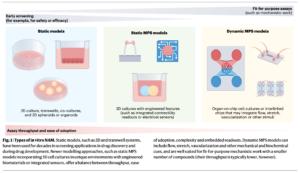
Categories of in vitro NAMs from Stresser, 2023.
Addressing key elements of technology readiness for in vitro NAMs will help get these tools out of research and into the hands of users. Components of readiness that are important to address at an early stage include assay performance, complexity of the protocol, cell sources, operations processes, and establishing the application domain.
My team is interested in the application of in vitro NAMs for the characterization of existing drugs and new drug candidates — to understand how drugs work and improve our knowledge about disease biology. Makingin vitro NAMs “FAIR” (findable, accessible, interpretable, and reusable) is a challenge we have been working on (see here).
I have spent my entire career on biological model systems (in vitro, ex vivo and in vivo) and assay development, throughout my time in academia, biopharma, and contract research. In shepherding the BioMAP platform of human primary cell and co-culture-based assays through concept to commercialization, I’ve learned some hard lessons about what it takes to get cell-based systems production ready.
As a strong advocate for in vitro NAMs I’m often asked to review the technology readiness of new platforms. I’m a biologist with drug discovery research experience, so view new methods from that perspective.
Below are some practical considerations I look for when assessing technology readiness.
Assay Performance
Assay performance assessments give us the key metrics for judging technology readiness. And yet there has been little alignment on standards for reporting basic performance metrics for in vitro NAMs. Separate guidelines have been developed for assessment of high throughput screening assays (Iversen, 2012), MPS platforms (Schurdak, 2020), and formal validation of NAMs for replacement of animal tests (Bal-Price, 2018, van der Zalm, 2022, ICCVAM Validation Workgroup Report, 2024).
Aligning on a common, minimal set of metrics for reporting basic assay performance of in vitro NAMs would facilitate communication between NAMs developers and help potential users gain confidence in these technologies. A common standard would also support integration of data across platforms. It is now clear that in vitro NAMs assays will be most useful, not in isolation, but together with other methods (along with biochemical and in silico NAMs) in combinatorial NAMs approaches (ACD Working Group Report, 2023).
A particularly useful set of data to have for evaluating the basic assay performance of any platform is from testing a single key reference agent (positive or active control) on a single key endpoint (anchor endpoint). The key reference agent should be tested in concentration response and with sufficient replicates to capture assay variability. Standard, basic assay metrics (%CV, Z-factor) derived from this data set provide important information about the readiness status of the platform. Such data also align with our expectations for animal models of efficacy. Using such data to track assay performance over time and drive assay optimization efforts also helps demonstrate commitment to assay quality and reproducible research.
Complexity of the Protocol
Complexity of the protocol can impact both scalability and reproducibility. Variations at individual steps accumulate to erode assay performance. Unnecessary steps increase variability of the final data. Overly complicated protocols also increase the likelihood that something will go wrong. With that view, have simpler options been tested? What data have been generated to support protocol decisions? Biologists often carry with us unconscious biases. Assay development teams are encouraged to consider protocol designs that that are as short and simple as practical.
Cell Sources
Establishing a reproducible source of cells or cell bank is critical for technology readiness (Bal-Price, 2018). Depending on the cell type (e.g., primary, iPSC-derived, transformed cell line, etc.), additional factors may be important. For example, if cells from single donors are used, how many donors have been tested? How were donors selected and is genetic characterization important? The use of pooled donors is an effective approach for establishing larger cell banks of primary cells if genetic characterization is not relevant for the context of use. How will cell quality will be determined and what cell quality acceptance criteria will be used (e.g. thresholds for % viability, biomarker expression, etc.) are important questions to have answered.
Operations Processes
Once an assay protocol has been defined and basic assay performance established, other elements, such as maturity of the laboratory and data management systems are considered. Is the protocol finalized (e.g., documented in a formal SOP or Standard Operating Procedure)? Are processes in place for experimental tracking and data management? Data management processes include documentation and tracking of experiments, reagents, samples, data sets, and data analysis methods. Attention to traceability and transparency also demonstrates an understanding of the risks to quality research and reproducible science.
Establishing the Application Domain
After performance characteristics have been defined and there is confidence that the assay or platform can produce trust-worthy results, experiments to establish the application domain can be reviewed. The application domain is defined by the types of materials that are compatible with the platform as well as the biological pathways and targets that can be detected. Technical limitations are identified through testing of diverse materials such as small and large molecules, cells, materials, mixtures, extracts, fluorescent compounds, and various vehicle formulations.
Establishing the biological scope of platform is accomplished by testing of reference agents with known mechanisms, including drugs, biologics, and chemical probes. Coverage of key drugs and mechanisms as well as testing at relevant concentrations are important. Testing of agents with nuisance mechanisms (Dahlin, 2021) as well as establishing counterscreens for cytotoxicity are needed to support accurate interpretation of assay results.
Other types of characterization (e.g., transcriptomics, proteomics profiling, single cell analysis, etc.) can also be helpful for understanding the biological scope of a particular in vitro NAM, and for comparing NAMs to one another. However, such data are not as impactful for technology readiness and establishing user confidence as are the data from direct testing of reference agents.
Summary
For researchers developing new in vitro NAMs of any type, addressing the considerations described above early in the process will help smooth the way to technology readiness.
The more quickly we can get in vitro NAMs platforms “technology ready” and into widespread use, the faster we will advance human relevant science for the benefit of patients.
If you have questions or need advice, get in touch.
Photo by Braden Collum on Unsplash.






